The Runaway model is an attempt to confront theory in sexual selection with demogenetic dynamics. The purpose of this module is to study the joint evolution of traits under sexual reproduction and of genetic architecture.
Quick description:
We model a theoretical population of individuals with genetically coded traits. These traits can be:
- the energy invested in gametes production (termed as “investment”),
- the energy invested in phenotype to signal quality (termed as “phenotype”),
- the energy invested in preference to be choosy on phenotype (termed as “preference”).
- the total resource is the sum of the above traits. It therefore affects further survival and logically the probability to participate in subsequent reproductions (termed as “resource”),
Each trait is coded by a number of diploid genes along the genome, and each gene has various effects on the traits. Through sexual selection and mutation, some variants are maintained, some removed and some created.
Although individuals can have a gender, reproduction uses sex: the amount of offspring an individual can have is dependent on its investment and the investment of its partner, and the genome of offspring is built through the meiosis principles. Sexual conflict is thus accounted for. One of the interest of the model is also to propose alternative mate choice routine and assess their impact: random mate choice, preference/phenotype based mate choice, intra-sexual competition.
Because the population is not subdivided in ecologically contrasted environment, there is no “local” adaptation peak. However, there might be some pseudo-equilibria conditional on all possible tactics in the population and on the genome structure.
A full pre-print paper describes the model assumptions, and provides some results as examples on BioRXiv
Installation
The model can be downloaded as a stand-alone version, including the core of the CAPSIS-4 platform, and the model code itself, here :
https://doi.org/10.15454/6NFGZ9
A new version will be made available shortly.
The model can be installed and executed on computers where the JAVA virtual machine is available. The Java VM can be downloaded here :
https://www.java.com/fr/download/
The user can launch the model using the capsis-en.bat (Windows) or the capsis-en.sh (Linux, MacOS) executables. More installation instructions can be found here: https://capsis.cirad.fr/capsis/documentation/installation_guide.
System requirements
As mentioned above, the Runaway model is portable to various OS as long as JAVA is installed. However, the model is agent-based, and creates potentially very large collections of objects (individuals, genomes).
It can therefore eat large quantities of memory, depending on user choices at the initialization, and on the length of simulations. A configuration using at least 16GB is advised for comfortable use. The model can make use of any amount of memory available in its 64bits version (i.e., if you use the 64bits version of JAVA). Multithreading is natively supported. Multiple instances of the model can also be launched simultaneously.
Several options are available to reduce memory usage:
- Reduce the carrying capacity of the populations simulated
- Record only a part of the time-steps in the simulation: Runaway indeed memorizes all steps, as a mean to explore each moment of the simulation in details, but this can be directly modified in the GUI (EXAMPLE COMING).
- Use RUNAWAY in script mode. To do this, please follow the guidelines here (LINK PAGE). You will find an example of script file to launch and record data in a simulation here (LINK TO SCRIPT FILE).
Quick start guide
At launch, start a project and select the RUNAWAY model:
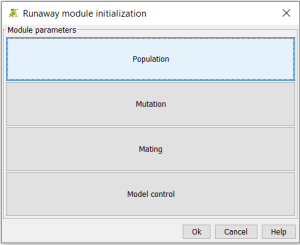
You will be prompted to an initialization dialogue box:
Each section links to parameters settings that the user can alter to simulate various scenario of evolution. The population panel allows to define the initial number of individuals, the carrying capacity parameter (that influence survival through density-dependence), the maximum number of loci involved in traits architecture, the autosome numbers and intra-autosomal recombination probabilities. Finally, the user can modify the total number of mutations than can be simulated per locus.
The mutation dialogue panel allows to modify the overall mutation rate. It also allows to influence the distribution of alleles effects for each trait in the simulation. The user can therefore plays with the two parameters of a beta distribution to generate a uniform mutation landscape (for instance, [1;1] settings), a bell-shaped distribution ([4;4]) or a skewed distribution (as in the default setting [25.4;0.65]).
The Mating dialogue box allows to simulate various mating systems. First, it allows to create subgroups for the reproduction that can be smaller than the whole population size. Second, it allows to link the fecundity to offspring number in two ways: either using the mean of gametic investment between parents (for instance mimicking the influence of direct benefits) or only the minimum of the two gametic investment (thereby representing the gametes number limitation). The encounter model allows to either let encounter occur at random within a mating group, or to rank it by using the competitiveness trait values. The preference model can be random (no preference), fixed threshold (rejection below, acceptation above), a probabilistic threshold, or unimodal preference. The model by default assumes mutual choice between individuals of both sexes, but it can be set to females choice only, a more “conventional sex roles” approach like. The span of the preference function can be absolute, meaning that it can encompass all the variation in the whole population, or it can on the opposite be expressed on the limited range of variation within the mating group. Preference is by default not directly costly, it therefore does not change the reproductive investment / survival trade-off; however the user can choose to model a scenario with direct cost on the preference. Another default assumption is that the population is composed of two sexes, but the model also allows to create sexual reproduction scenarios without referring to gender (as in paramecia for instance).
Finally, it is possible to include a sexual chromosome in the genome, allowing part of the loci to be restricted to one sex only.
The last dialogue box allows to tune the fecundity in the model, with the use of a demographic constant. By default, the value is set at 10. It means that if, for instance, the mean of gametic investment of two mated parents is 0.5, then the number of offspring will be the floor integer value 0.5*10 (here, 5). This constant allows to start simulation with trait values that would readily crash the population on the one hand, or to allow the irrealistic generation of too many individuals, which is memory and time consuming for the simulation. Another control variable is the maximum iteration of the mating loop: in some cases, when individuals try to choose each other, their preference may not easily produce matchings. This number simply avoid unending yet unfruitful dating.
Once the model initialized, you can start a first simulation by right-clicking on the first time step, choosing 'evolution' and setting your desired number of time-steps.
Once the simulation is finished, you can also do manual replicates, by repeating the same operation from the initial time step, or any new time step of interest.
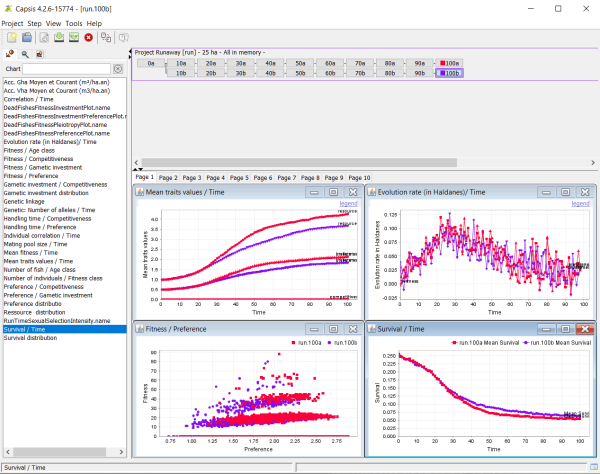
When the simulations are over, you can explore them using monitors, located on the left panel. To do so, click on a time step: it will set a colour square on it. Then click on a monitor, for instance, 'Mean traits values / Time'. The monitor will extract the required data from the simulation and display them. You can click once on another time step, for instance from a different simulation, and control+click on the monitor window: it will add these new data to the monitor, and you will thus be able to compare them. Note that you can also start a second project, using very different settings, and compare its results using the same framework on the same monitors. You can further explore the benefits of the CAPSIS-4 simulation platform using the contextual menus using right-clicking on figures → Configure → Help, or by using the contextual help buttons on dialogue boxes.
Post-Scriptum:
The interface, monitors, as well as some options of the model are in regular development. If you require a stable version, please contact us. If you require the code of the model, or would like to collaborate on some possible modifications of the model, you can also contact us.