Table of Contents
Data Extractor
[To be reviewed and clarified - fc - 8.12.2010]
Presentation
Data Extractors are among the most used Capsis extensions. These tools extract data in Capsis projects in order to draw various diagrams, tables or other representations to check what happens during the simulations. An extractor is synchronized on a project target Step, carrying a situation at a given date. It generally shows either the state at this date, e.g. an histogram, or the state evolution in the scenario from the root step to this target step, e.g. a curve.
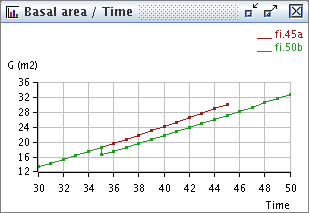
Several extractors can be added in a single window (a data block) to be rendered in the same graphic. This way, several curves for various scenarios can be compared easily (fig. 1). The resulting graphic is rendered by an other extension (Dara Renderer). A graph can be renderer in different way e.g. curves, histograms, scatterplots, tables. Use Right click on the graph to see available renderer.
Fig. 1. A Data Block with two Data Extractors “Basal area over Time” synchronized on two steps “45a” and “50b” in project named “fi”, represented by the “DRCurves” Data Renderer
Write your own extractor
Common extractor types
We distinguish 3 type of Data Extractor
- Data over Time : DETimeX
- Multi Data : DEMultiX
- Multi Data over time : DEMultiTimeX
Some function must be overriden for each type of extractor
Create a Data Extractor
- Create the class in
“mymodel”/extension/dataextractor - Create translation files
“mymodel”/extension/dataextractor/Labels_en.properties“mymodel”/extension/dataextractor/Labels_fr.properties
Translation file
The translation file should contains the labels of the extractors
For instance template/extension/dataextractor/Labels_en.properties:
DETimeN=N / Time DETimeN.yLabel=N DETimeN.description=N / Time DESpeciesN=N / Species DESpeciesN.yLabel=N DESpeciesN.description=N / Species DESpeciesTimeN=N / Time (by Species) DESpeciesTimeN.yLabel=N DESpeciesTimeN.description=N / Time (by Species)
Note : new extractor doesn't need to load explicitly translation file in a static block.
Data over time (DETimeX)
/** * @author __AUTHOR__ __DATE__ */ public class DETimeN extends DETimeX { public static final String AUTHOR = "__AUTHOR__"; public static final String VERSION = "1.0"; static public boolean matchWith(Object referent) { if (!(referent instanceof TplModel)) {return false;} return true; } /** return a value for a type and a step */ @Override protected Number getValue(GModel m, GScene stand, int date) { Collection<Tree> trees = doFilter (stand); return trees.size(); } }
Multi Data (DEMultiX)
/** * @author __AUTHOR__ __DATE__ */ public class DESpeciesN extends DEMultiX { public static final String AUTHOR = "__AUTHOR__"; public static final String VERSION = "1.0"; static public boolean matchWith(Object referent) { if (!(referent instanceof TplModel)) {return false;} return true; } /** return a value for a type and a step */ @Override protected Number getValue(GModel m, GScene stand, int classId, Object cl) { Collection<Tree> trees = doFilter (stand); int n = 0; for(Tree t : trees) { TplTree tt = (TplTree) t; if(tt.getSpecies() == cl) { n++; } } return n; } /** Return a list of type of curve */ @Override protected List<Object> getTypes(GScene stand) { List<Object> l = new ArrayList<Object>(); l.add(TplSpecies.BEECH); l.add(TplSpecies.OAK); return l; } }
Multi Data over time (DEMultiTimeX)
idem as previous, by for each step.
public class DESpeciesTimeN extends DEMultiTimeX { public static final String AUTHOR = "__AUTHOR__"; public static final String VERSION = "1.0"; static public boolean matchWith(Object referent) { if (!(referent instanceof TplModel)) {return false;} return true; } /** return a value for a type and a step */ @Override protected Number getValue(GModel m, GScene stand, int classId, Object cl) { Collection<Tree> trees = doFilter (stand); int n = 0; for(Tree t : trees) { TplTree tt = (TplTree) t; if(tt.getSpecies() == cl) { n++; } } return n; } /** Return a list of type of curve */ @Override protected List<Object> getTypes(GScene stand) { List<Object> l = new ArrayList<Object>(); l.add(TplSpecies.BEECH); l.add(TplSpecies.OAK); return l; } }
Specific extractor
For more specific extractor, we can inherit directory from AbstractDataExtractor. In this case, we must implements more methods.